Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
Por um escritor misterioso
Last updated 10 novembro 2024


Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
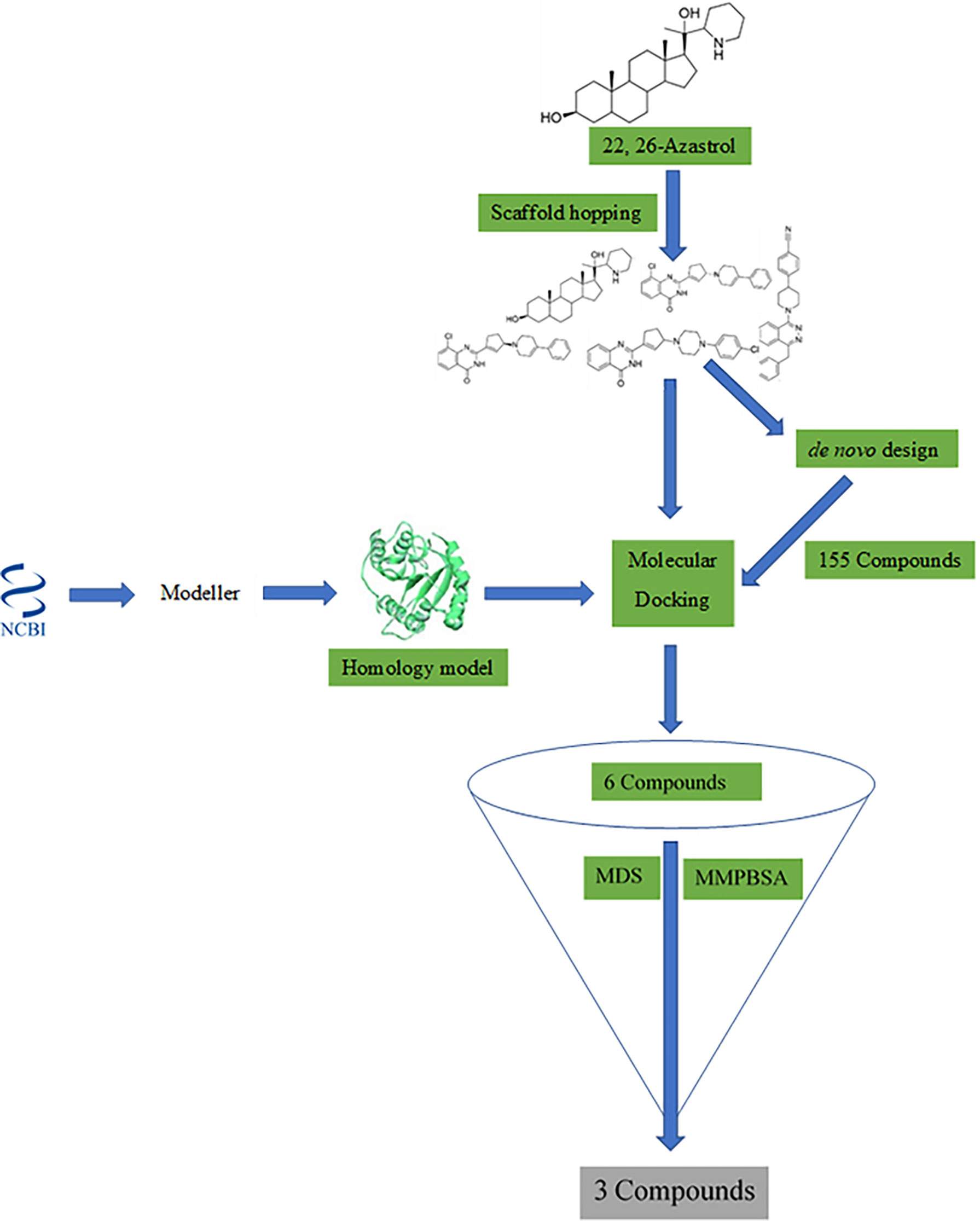
Frontiers Homology Modeling, de Novo Design of Ligands, and Molecular Docking Identify Potential Inhibitors of Leishmania donovani 24-Sterol Methyltransferase
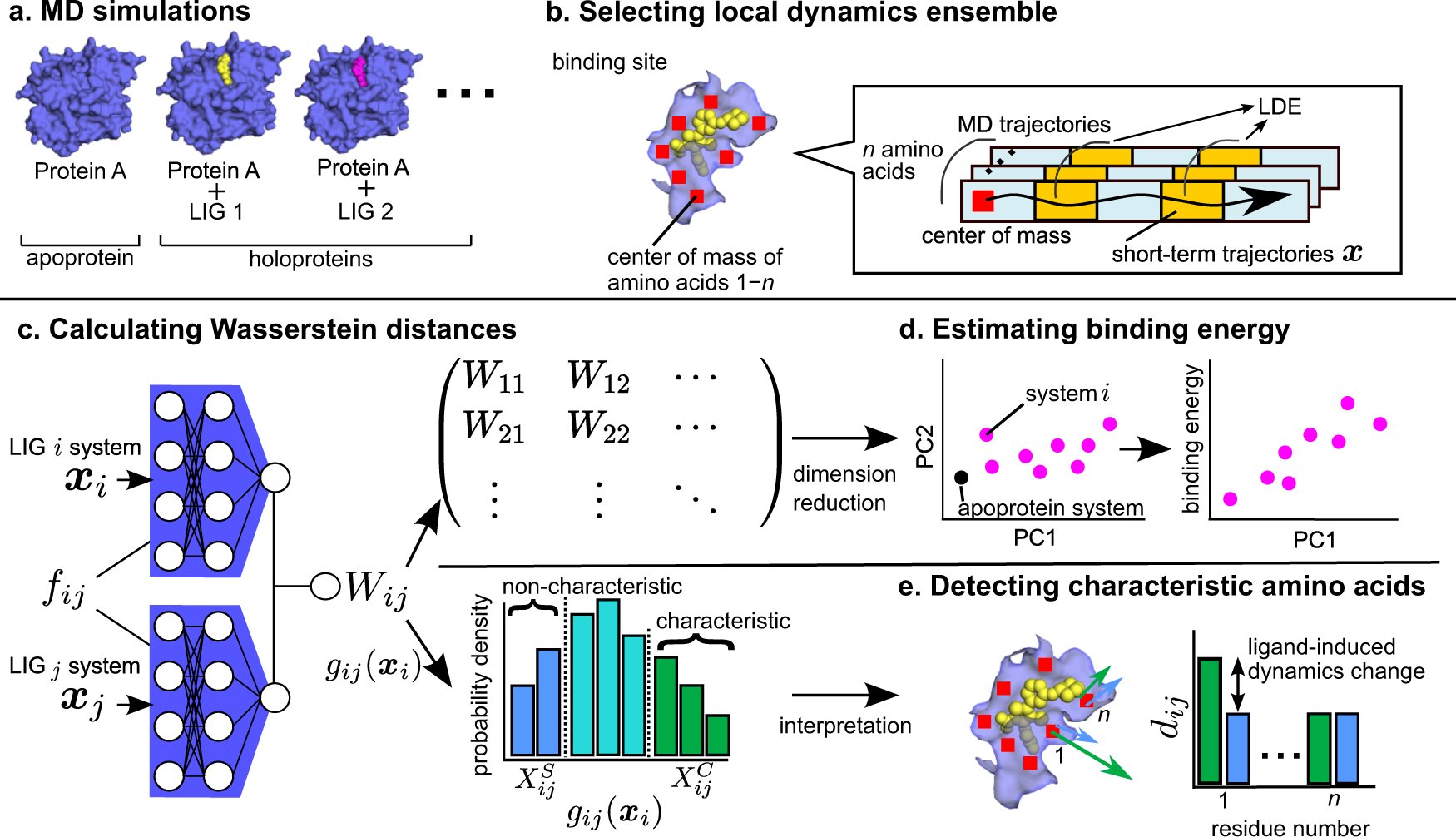
Differences in ligand-induced protein dynamics extracted from an unsupervised deep learning approach correlate with protein–ligand binding affinities
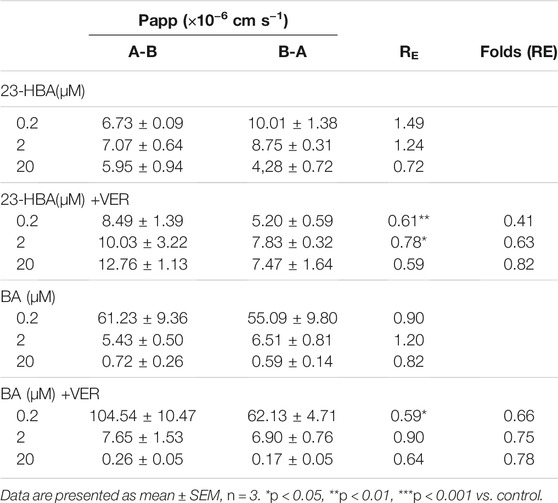
Frontiers Involvement of P-gp on Reversing Multidrug Resistance Effects of 23-Hydroxybetulinic Acid on Chemotherapeutic Agents

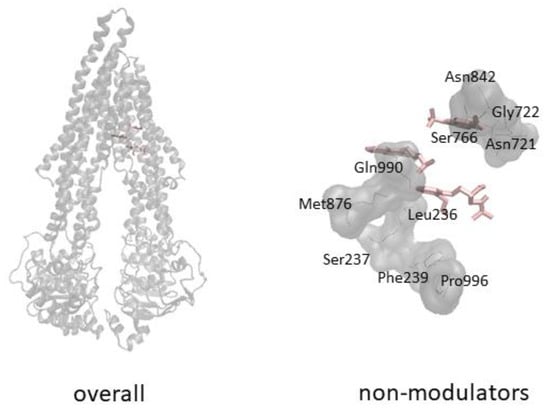
Structures of P-glycoprotein in different conformations (A) Domain

Machine learning approaches and their applications in drug discovery and design - Priya - 2022 - Chemical Biology & Drug Design - Wiley Online Library
Cells, Free Full-Text
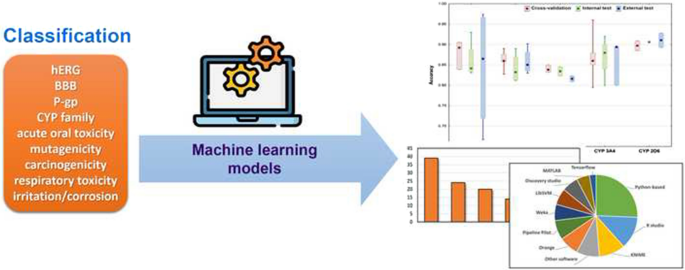
Machine learning models for classification tasks related to drug safety
Recomendado para você
-
 Brain Test Level 371, 372, 373, 374, 375, 376, 377, 378, 379, 380 Answers10 novembro 2024
Brain Test Level 371, 372, 373, 374, 375, 376, 377, 378, 379, 380 Answers10 novembro 2024 -
 Brain Test Level 202 Solve the puzzle in 202310 novembro 2024
Brain Test Level 202 Solve the puzzle in 202310 novembro 2024 -
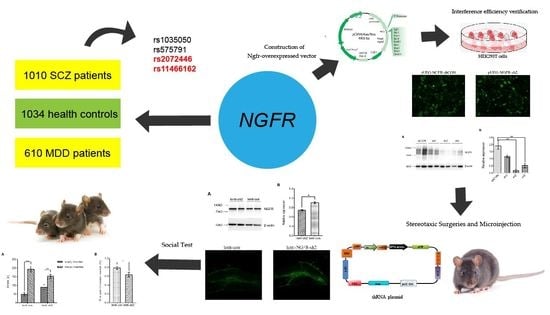 Brain Sciences, Free Full-Text10 novembro 2024
Brain Sciences, Free Full-Text10 novembro 2024 -
Optical challenge: A Sherlock Holmes-like mind can find the hidden potato in 11 seconds! - alro news10 novembro 2024
-
 BRAIN TEST Poster for Sale by HMS STORE10 novembro 2024
BRAIN TEST Poster for Sale by HMS STORE10 novembro 2024 -
 brain test 4 level 24110 novembro 2024
brain test 4 level 24110 novembro 2024 -
Imaging Surrogates of Disease Activity in Neuromyelitis Optica Allow Distinction from Multiple Sclerosis10 novembro 2024
-
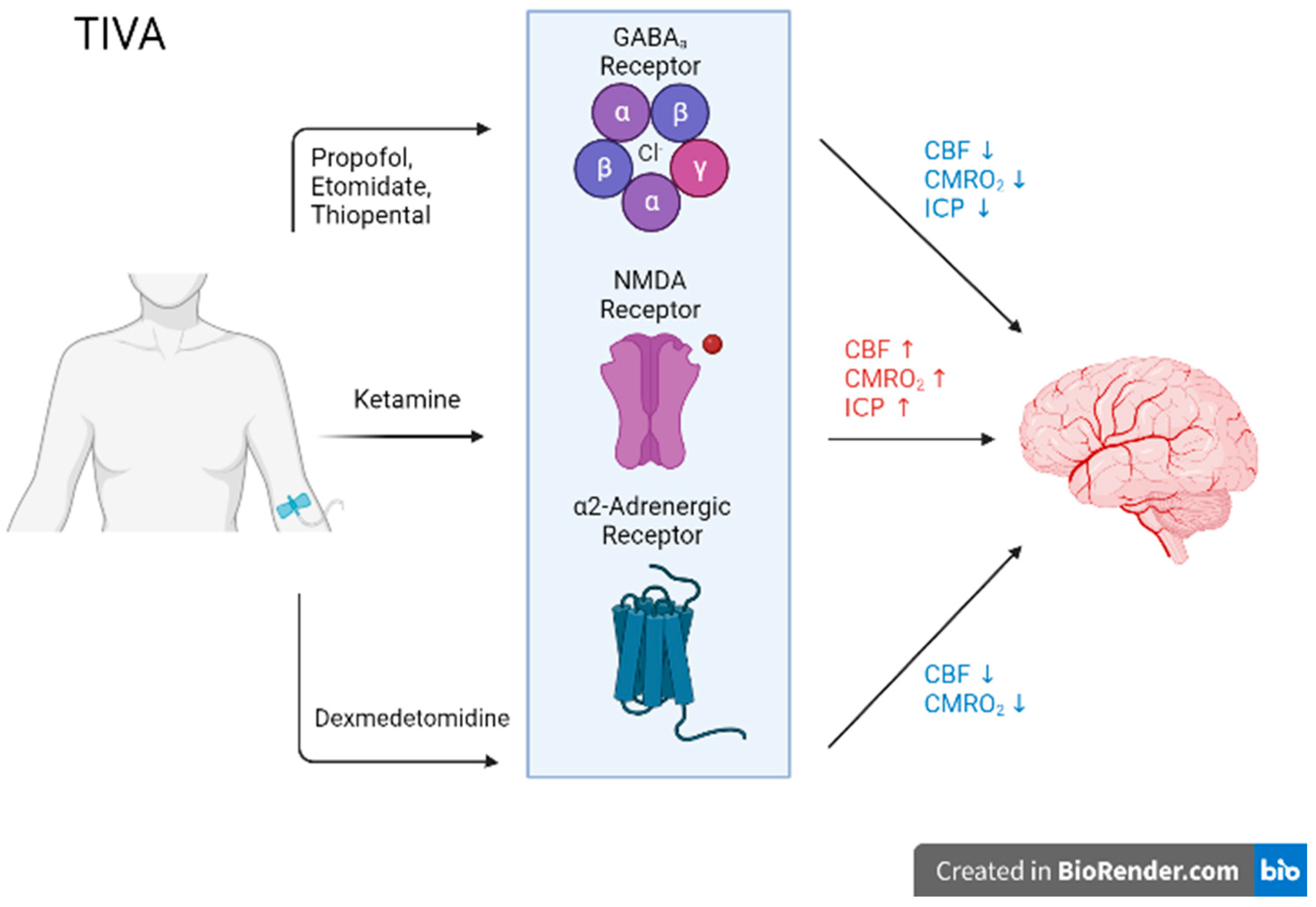 Biomedicines, Free Full-Text10 novembro 2024
Biomedicines, Free Full-Text10 novembro 2024 -
MTVBaseAfrica10 novembro 2024
-
 Think about your thoughts #stereo10 novembro 2024
Think about your thoughts #stereo10 novembro 2024
você pode gostar
-
 Zuma Dubai - FACT Magazine10 novembro 2024
Zuma Dubai - FACT Magazine10 novembro 2024 -
 Assista ao comparativo gráfico de The Legend of Zelda: Twilight10 novembro 2024
Assista ao comparativo gráfico de The Legend of Zelda: Twilight10 novembro 2024 -
 Nintendo DS - ROM set (ROM set under construction10 novembro 2024
Nintendo DS - ROM set (ROM set under construction10 novembro 2024 -
 diario de um vampiro gifs10 novembro 2024
diario de um vampiro gifs10 novembro 2024 -
 osfilmes™ on X: Drive do filme: Five Nights at Freddy's - O10 novembro 2024
osfilmes™ on X: Drive do filme: Five Nights at Freddy's - O10 novembro 2024 -
 Cyberpunk Sci-Fi Night City HD 4K Wallpaper #8.144510 novembro 2024
Cyberpunk Sci-Fi Night City HD 4K Wallpaper #8.144510 novembro 2024 -
 SANRIO COLLECTION, Kawaii pajamas and underwears10 novembro 2024
SANRIO COLLECTION, Kawaii pajamas and underwears10 novembro 2024 -
Pokémon Liga Índigo: 1ª Temporada, Episódio 3, Liga Índigo: Ash Pega um Pokémon! (Episódio 3), By Project Animes10 novembro 2024
-
 ⚡Pikamee shows her New Intros & Backgrounds10 novembro 2024
⚡Pikamee shows her New Intros & Backgrounds10 novembro 2024 -
 Segundo a VGC, Far Cry 7 levará a franquia para o lado totalmente online10 novembro 2024
Segundo a VGC, Far Cry 7 levará a franquia para o lado totalmente online10 novembro 2024


